MoBIE
MultiModal Big Image Data Sharing and Exploration
Volume registration
This tutorial will show how to generate a project from multimodal volume images and register them.
retrieve the raw data
-
To download the Electron microscopy (EM) dataset, head to https://www.ebi.ac.uk/empiar/EMPIAR-11537/, make sure to de-select all data except #4 “Downscaled (20nm) aligned FIB SEM stack resliced to match the Airyscan dataset” and click the individual download button (choose HTTP).
-
The Fluorescence microscopy (FM) dataset is stored at https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BSST1075. You can download the raw data file directly from here.
create the MoBIE project
- open the LM data in Fiji using the BioFormats Importer plugin. (
Plugins > Bio-Formats > Bio-Formats Importer) - you should get a Hyperstack image with 4 channels. (MitoTracker, Agglutinin, GFP-TGN46, and Hoechst)
- adjust the lookup table for each channel to fit the fluorophore. (
Image > Lookup Tables) - Currently, the MoBIE project creator only supports single channels, so we have to split the fluorescence stack into separate channels using
Image > Color > Split Channels.
-
create a new MoBIE project: type “mobie” in the search bar or choose
Plugins > MoBIE > Create > Create New MoBIE project...
name the project as you like and create it in a local directory.
- add a new dataset, name it as you like. Do not tick “2D”.
- select the first channel image in Fiji
- add a new source
current displayed image, call it appropriately (channels are sorted inverse), make sure the Image type is “Image” and do not tickmake view exclusive.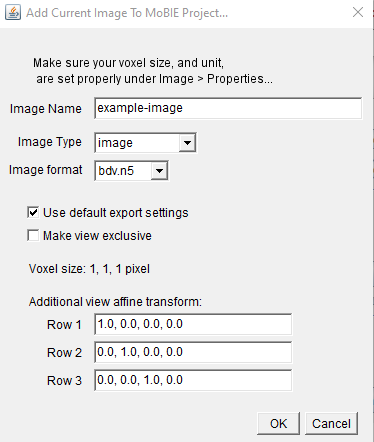
-
choose the
selection group nameas “fluorescence” or “FM”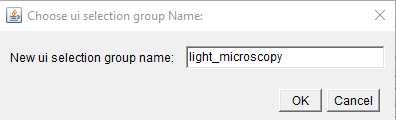
- open the MoBIE project to see how the scaling and LUT are transferred
- continue the same way with the other channels, adding them to the same dataset and selection group.
- open the other channel views using the MoBIE UI and explore the multi-channel volume
- right click into the multi-channel viewer and
Save current view Save as new view, save toprojcect- Call the view something like “all channels” and make it part of the “FM” group
- close MoBIE
- open the EM data in Fiji
- add the volume to MoBIE under the selection group “EM” (make new selection group) to the same dataset. The format conversion can take a couple of minutes.
- open the MoBIE project and view your multichannel FM and EM together
- make all fluorescence channels but the Hoechst invisible by clicking on
S - change contrast and transparency settings for the relevant sources (
B)
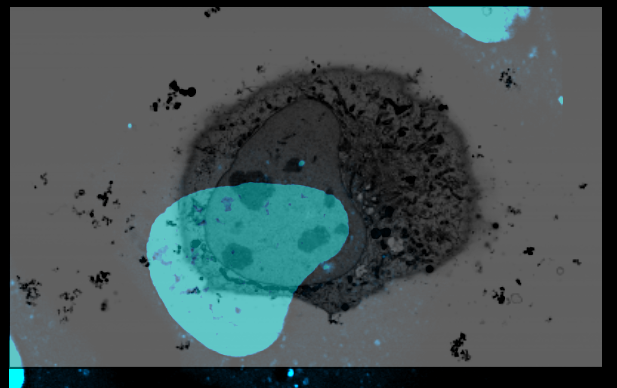
Manual registration of sources
- right click the image “registration manual”, select the EM image as we want to keep the multi-channels where they are.
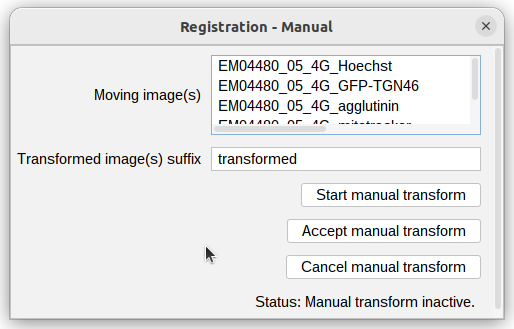
- Click
Start manual transform - use the right mouse button to drag the image around.
- Press the
zkey to make sure rotations are around the viewing axis and use the right and left arrow keys to rotate. - You can also use the mouse wheel to translate in
z, and the up and down arrows to scale (dangerous). - If necessary adjust brightness or opacity of the images using MoBIE’s main window (
B). - click
cancel manual Transformto undo the translation and bring the image back to its original position. - click
Accept manual Transformto make it permanent and store the view into the project. This will only save the transformed EM source. Create a new selection group “registered”. - clear the viewer
- open the registered view an all FM channels. Check their registration by scrolling through the volume.
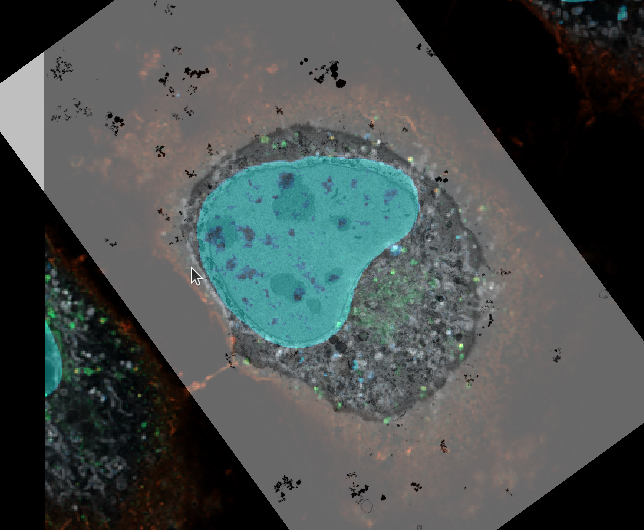
- push
CTRL + yto view the volume from the side. Make sure te mouse pointer is in the center of the feature, as it determines the viewer’s rotation axis. - repeat the manual registration of the EM volume until your registration matches in all axes.
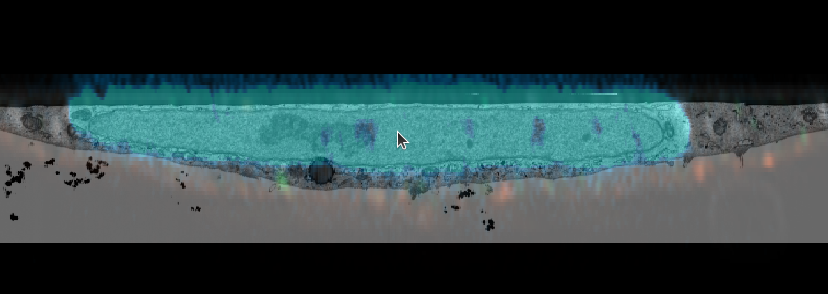
- You could also use the mitochondria or another channel for refining it.
Note that when clicking Accept manual transform the single transformed source will be saved as a new view. In case you like to transform more.
- To save a complete view with all sources (channels + EM), do
right click > save current viewand save your view as desired.
(optional) registration using BigWarp
You might realize, that a rigid transform (composed of translations, rotations and isotropic scaling) is not sufficient to register all regions of the volumes at once. In order to achieve this, we need to allow for more flexibility. We can apply a landmark-based registration and use BigWarp for this purpose. A good introduction in the concepts and functions can be found here.
right click > Registration - BigWarp- you can select multiple sources as the moving image, in this case, select all fluorescence channels. (You should change LUT, contrast and opacity settings for the relevant channel(s) to make it easier to identify objects as landmarks).
- when you start BigWarp, multiple windows will open. One showing the fixed image (EM), one showing the moving image (FM) and a list of landmarks.
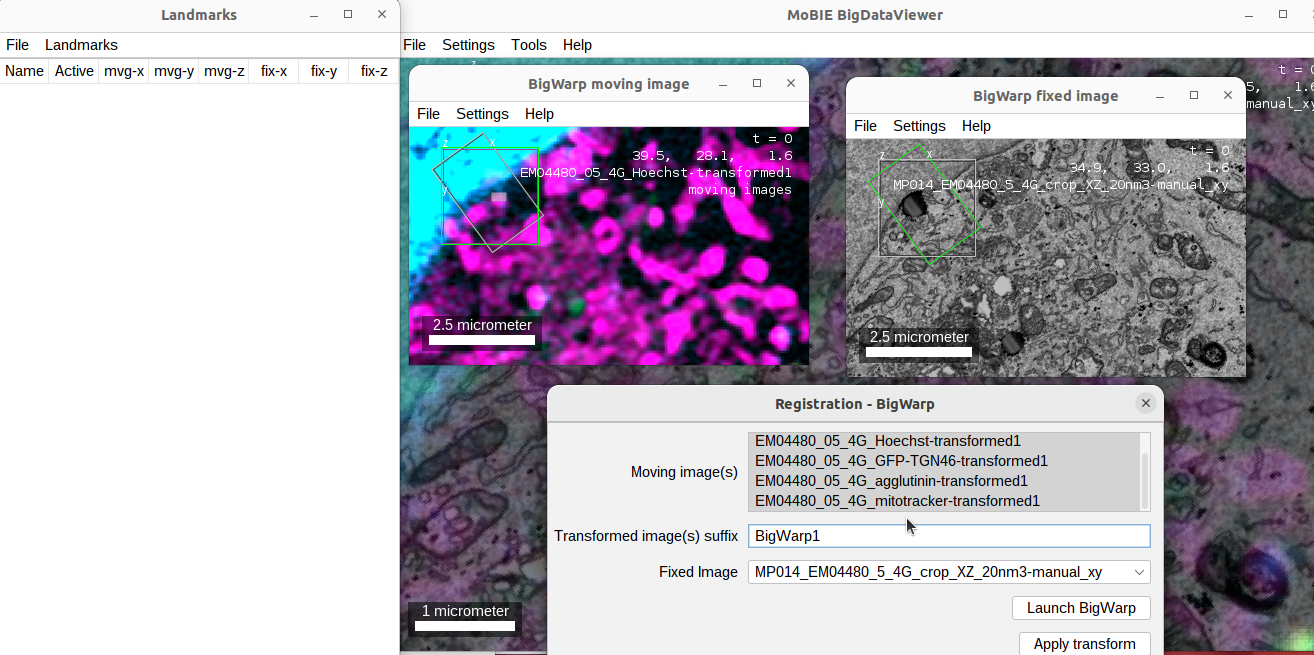
- it is very useful to open the help which gives you an overview of all inputs and keyboard shortcuts.
- pressing
Fwill switch between single-source and fused mode showing the moving and fixed source(s) in one viewer. - one very useful one is pressing
Tin the fixed image viewer to toggle a fused view of all sources on or off. - to start placing landmarks, we have to activate BigWarp’s landmark mode by pressing
␣ Space. - now add corresponding landmarks by clicking in both image viewers
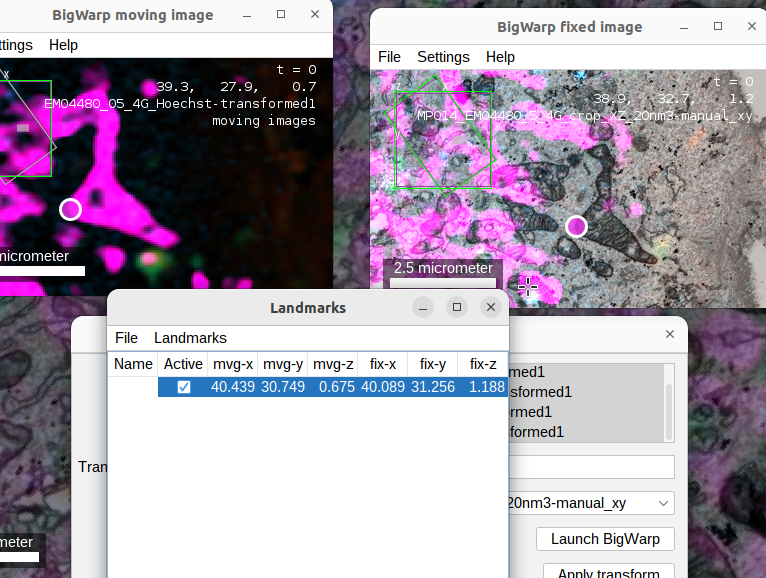
- if you want to browse a viewer (using the standard commands) switch off landmark mode again.
- when you added a couple of landmarks, you can toggle a transformed view by pressing
T.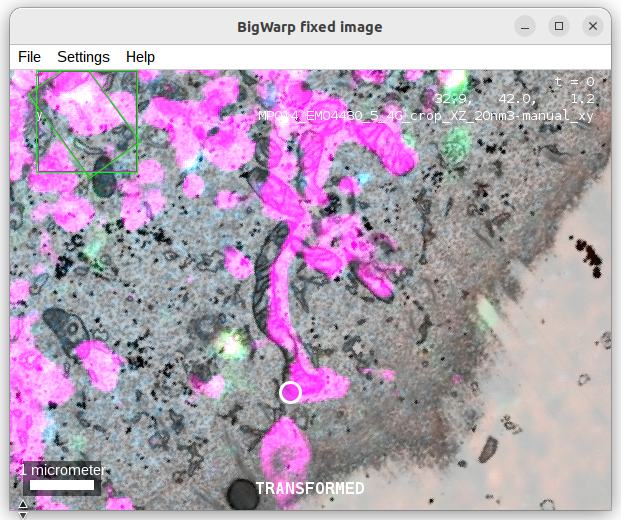
- Observe how the registration improves with adding more landmarks.
- press
Uto open the BigWarp preferences. -
observe how the registration changes/improves when selecting a different transformation type (“affine” vs. “similarity” etc.).
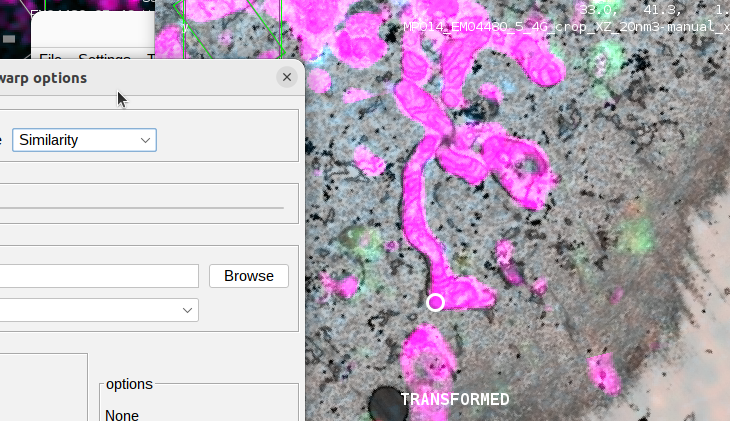
- if you are interested in comparing your registration with the published result, you can open BigWarp using the raw (not transformed) sources (FM as moving sources) in BigWarp and apply the transformations using the published landmarks that you can download here. Click
File > Import landmarksto load them into BigWarp.