MoBIE
MultiModal Big Image Data Sharing and Exploration
Correlative light and electron microscopy
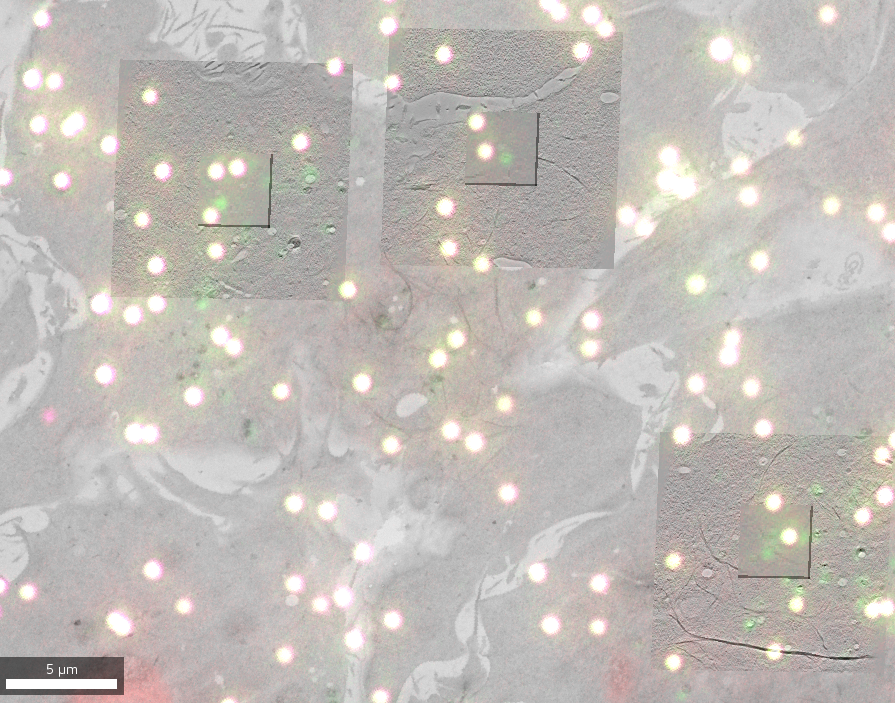
A correlative light and electron microscopy (CLEM) project combines imaging data from both modalities to benefit from the dynamic and specific localization of labeled entities (Fluorescence Microscopy, FM) and the high resolution structural and contextual information (Electron Microscopy, EM).
Data & project set-up
The data for this project comes from two different experiments (one using S. cerevisiae, one using a HeLa cell line) that are available as different datasets. It contains 2D FM images to target features of interest and EM data in both 2D (overview maps) and 3D (reconstructed electron tomographic volumes at different resolutions).
The original data in various formats (MRC, TIF, etc.) was converted to the MoBIE data format using the MoBIE python library.
Initial registrations were imported using the metadata from SerialEM and ec-CLEM software and then refined using MoBIE’s transformation functionality.
Exploring the project
Open the project from https://github.com/mobie/clem-example-project. See “Getting Started” for how to open a project in the MoBIE Fiji plugin. The project will open the default view, which shows the lowest magnification overview EM map.
You can then add any other source (whether EM or fluorescence) to it or choose one of the pre-defined detail_area views that contain all local sources.